Regularization in Neural Networks¶
Table of Contents¶
@[toc]
Introduction¶
If the training dataset is not large enough, due to the high flexibility and capacity of deep learning models, overfitting can be a severe problem. To address this, regularization methods are introduced. In neural networks, besides adding regularization functions in activation layers, dropout also serves a regularization purpose. Let’s explore this.
Prerequisites¶
Before starting, we need to import the required libraries and load the data. Some dependencies can be downloaded from here.
# coding=utf-8
import matplotlib.pyplot as plt
from reg_utils import compute_cost, predict, forward_propagation, backward_propagation, update_parameters
from reg_utils import sigmoid, relu, initialize_parameters, load_2D_dataset
from testCases import *
# Load dataset
train_X, train_Y, test_X, test_Y = load_2D_dataset()
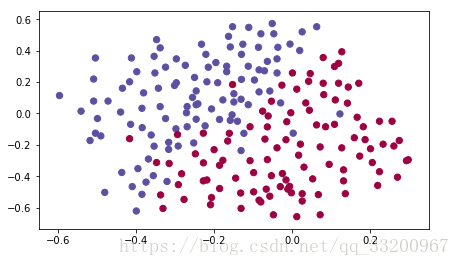
The dataset used in this example is visualized as follows:
Model Function¶
We’ll implement a model function to test and compare three scenarios:
1. Without regularization
2. With L2-regularized activation function
3. With dropout
def model(X, Y, learning_rate=0.3, num_iterations=30000, print_cost=True, lambd=0, keep_prob=1):
"""
Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.
Arguments:
X -- Input data, shape (input size, number of examples)
Y -- True "label" vector (red:1, blue:0), shape (output size, number of examples)
learning_rate -- Learning rate for optimization
num_iterations -- Number of iterations in optimization loop
print_cost -- If True, print cost every 10000 iterations
lambd -- Regularization hyperparameter (scalar)
keep_prob -- Probability of keeping neurons active during dropout
Returns:
parameters -- Parameters learned by the model, used for prediction
"""
grads = {}
costs = [] # Track cost
m = X.shape[1] # Number of examples
layers_dims = [X.shape[0], 20, 3, 1]
# Initialize parameters
parameters = initialize_parameters(layers_dims)
# Gradient descent loop
for i in range(num_iterations):
# Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID
if keep_prob == 1:
a3, cache = forward_propagation(X, parameters)
elif keep_prob < 1:
a3, cache = forward_propagation_with_dropout(X, parameters, keep_prob)
# Cost function
if lambd == 0:
cost = compute_cost(a3, Y)
else:
cost = compute_cost_with_regularization(a3, Y, parameters, lambd)
# Backward propagation
assert (lambd == 0 or keep_prob == 1) # Can use L2 and dropout together, but this task explores one at a time
if lambd == 0 and keep_prob == 1:
grads = backward_propagation(X, Y, cache)
elif lambd != 0:
grads = backward_propagation_with_regularization(X, Y, cache, lambd)
elif keep_prob < 1:
grads = backward_propagation_with_dropout(X, Y, cache, keep_prob)
# Update parameters
parameters = update_parameters(parameters, grads, learning_rate)
# Print cost every 10000 iterations
if print_cost and i % 10000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
if print_cost and i % 1000 == 0:
costs.append(cost)
# Plot cost
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (x1,000)')
plt.title("Learning rate = " + str(learning_rate))
plt.show()
return parameters
Without Regularization¶
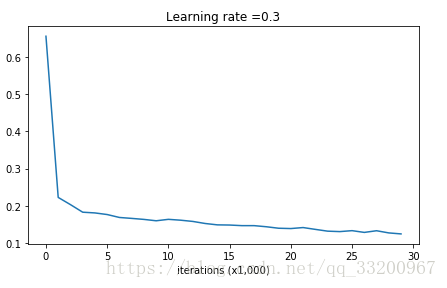
Run the model without regularization to observe overfitting:
if __name__ == "__main__":
parameters = model(train_X, train_Y)
print("On the training set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
Output:
Cost after iteration 0: 0.6557412523481002
Cost after iteration 10000: 0.16329987525724216
Cost after iteration 20000: 0.13851642423255986
On the training set:
Accuracy: 0.947867298578
On the test set:
Accuracy: 0.915
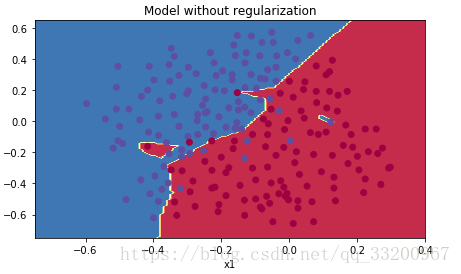
The cost curve shows overfitting:
L2 Regularization on Activation Functions¶
Loss Function¶
For L2 regularization, we modify the loss function:
Standard cross-entropy cost:
\(\(J = -\frac{1}{m} \sum\limits_{i = 1}^{m} \left( y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \right) \tag{1}\)\)
Regularized cross-entropy cost:
\(\(J_{regularized} = \underbrace{-\frac{1}{m} \sum\limits_{i = 1}^{m} \left( y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \right)}_{\text{cross-entropy cost}} + \underbrace{\frac{1}{m} \frac{\lambda}{2} \sum\limits_l\sum\limits_k\sum\limits_j W_{k,j}^{[l]2}}_{\text{L2 regularization cost}} \tag{2}\)\)
Implementation Code¶
def compute_cost_with_regularization(A3, Y, parameters, lambd):
"""
Implements the cost function with L2 regularization.
Arguments:
A3 -- Post-activation output of forward propagation, shape (output size, number of examples)
Y -- True label vector, shape (output size, number of examples)
parameters -- Python dictionary containing parameters W1, W2, W3
Returns:
cost -- Regularized cost value
"""
m = Y.shape[1]
W1 = parameters["W1"]
W2 = parameters["W2"]
W3 = parameters["W3"]
cross_entropy_cost = compute_cost(A3, Y) # Cross-entropy cost
L2_regularization_cost = (1 / m) * (lambd / 2) * (
np.sum(np.square(W1)) + np.sum(np.square(W2)) + np.sum(np.square(W3)))
cost = cross_entropy_cost + L2_regularization_cost
return cost
Backward Propagation with Regularization¶
def backward_propagation_with_regularization(X, Y, cache, lambd):
"""
Implements backward propagation with L2 regularization.
Arguments:
X -- Input dataset, shape (input size, number of examples)
Y -- True label vector
cache -- Output from forward_propagation()
lambd -- Regularization hyperparameter
Returns:
gradients -- Dictionary of gradients
"""
m = X.shape[1]
(Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1. / m * np.dot(dZ3, A2.T) + (lambd / m) * W3
db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1. / m * np.dot(dZ2, A1.T) + (lambd / m) * W2
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1. / m * np.dot(dZ1, X.T) + (lambd / m) * W1
db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3, "dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
Run L2 Regularized Model¶
if __name__ == "__main__":
parameters = model(train_X, train_Y, lambd=0.7)
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
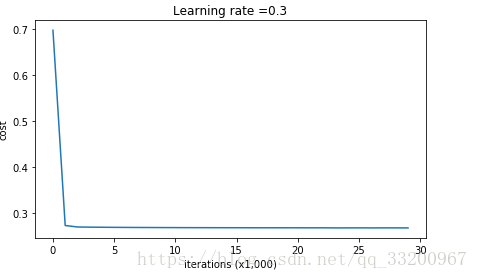
Output:
Cost after iteration 0: 0.6974484493131264
Cost after iteration 10000: 0.2684918873282239
Cost after iteration 20000: 0.2680916337127301
On the train set:
Accuracy: 0.938388625592
On the test set:
Accuracy: 0.93
Cost curve:
Convergence plot:
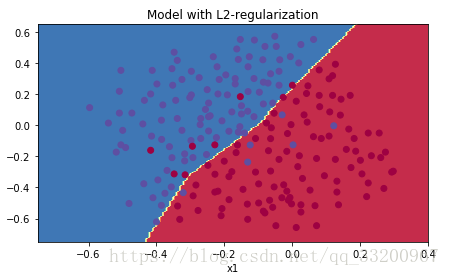
What L2 Regularization Does¶
L2 regularization assumes simpler models have smaller weights. By penalizing weight squares, we drive weights toward smaller values, resulting in smoother models where output changes more slowly with input changes.
Dropout¶
Dropout is a specialized regularization technique for deep learning that randomly deactivates neurons during training.
Dropout Process¶
During forward propagation, some neurons are randomly turned off (masked to 0). During backward propagation, the same neurons are ignored.
Visualization:
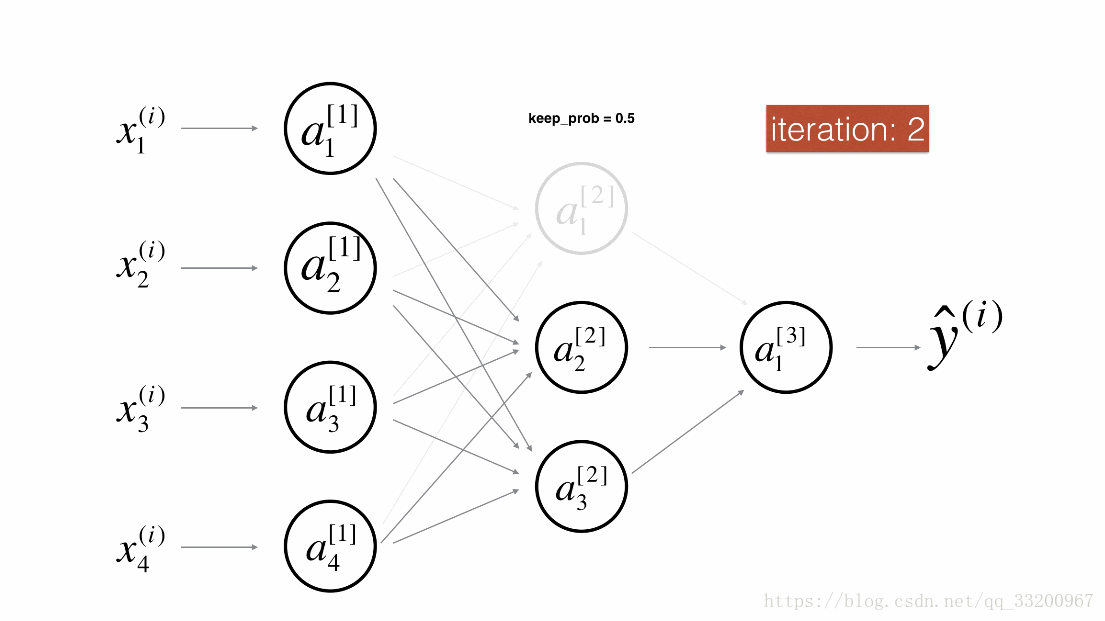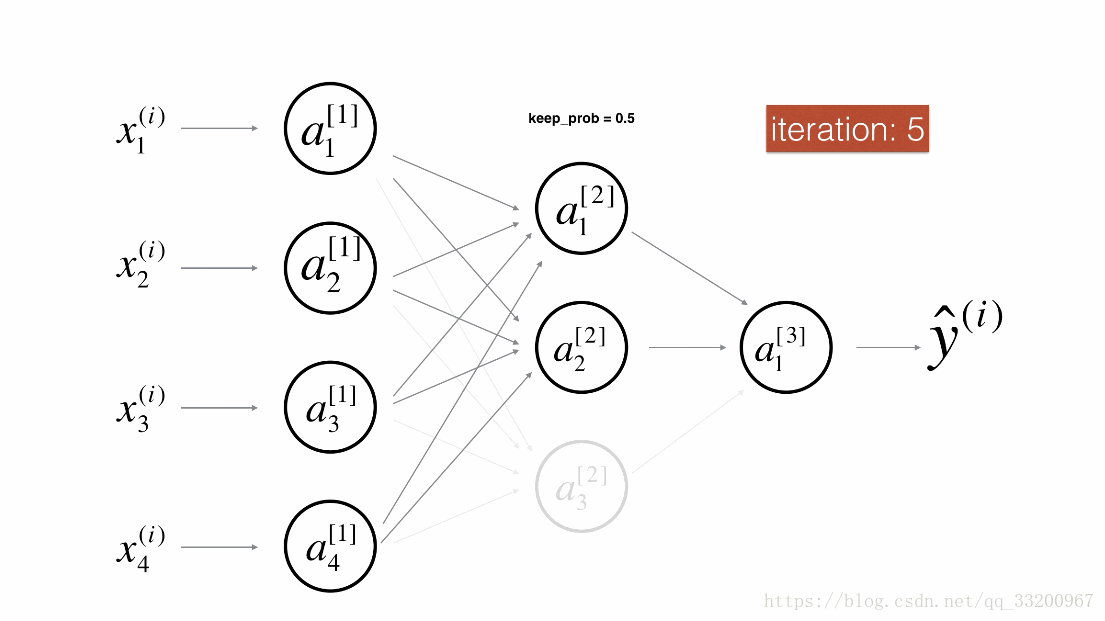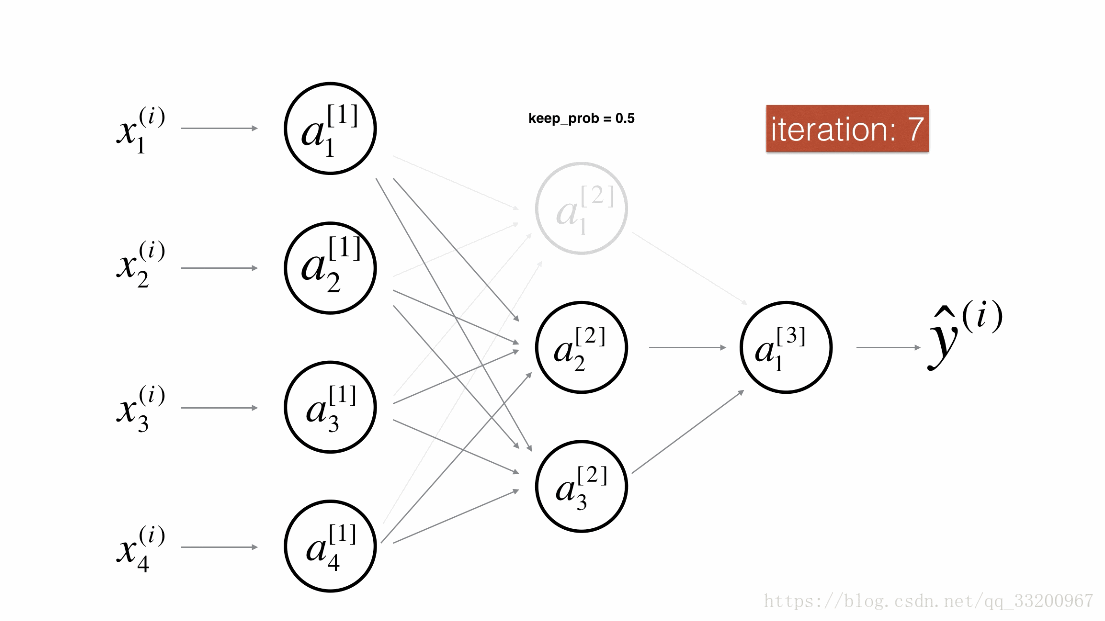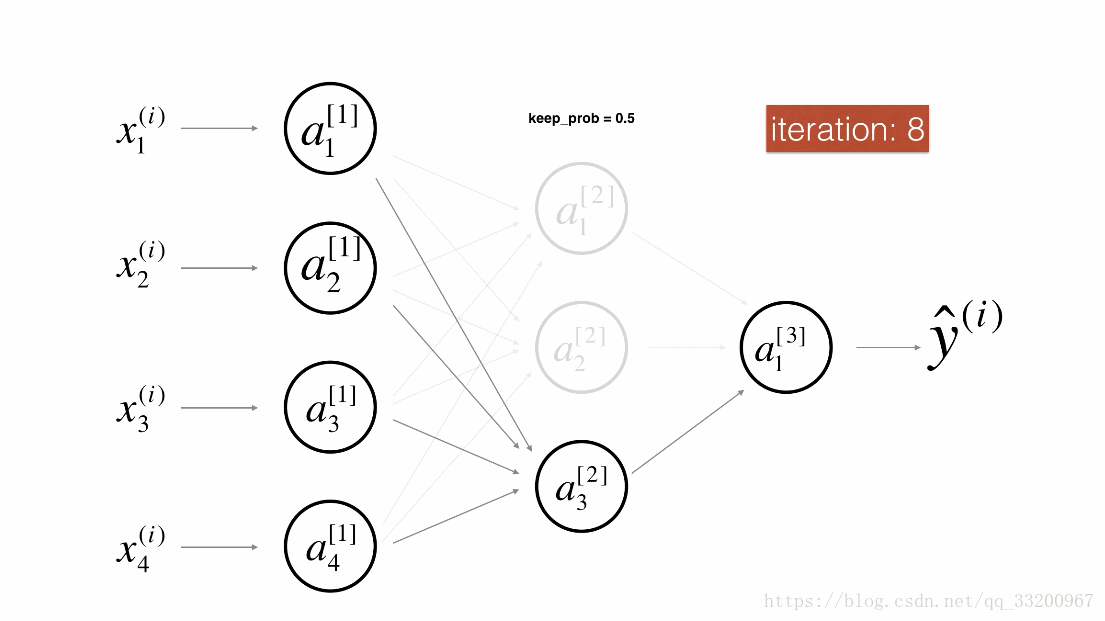
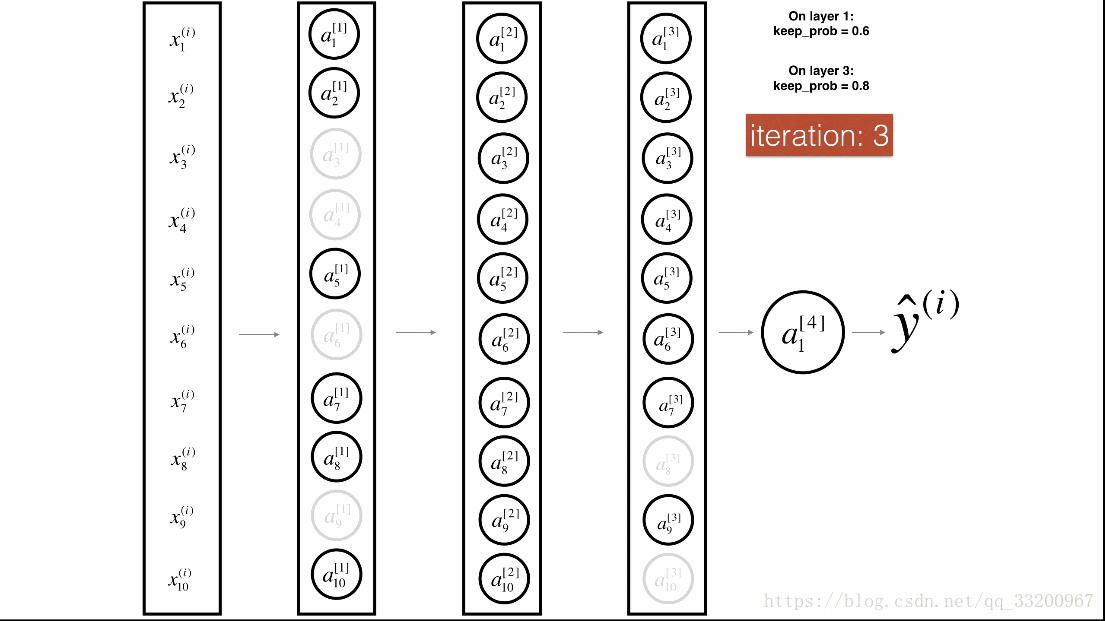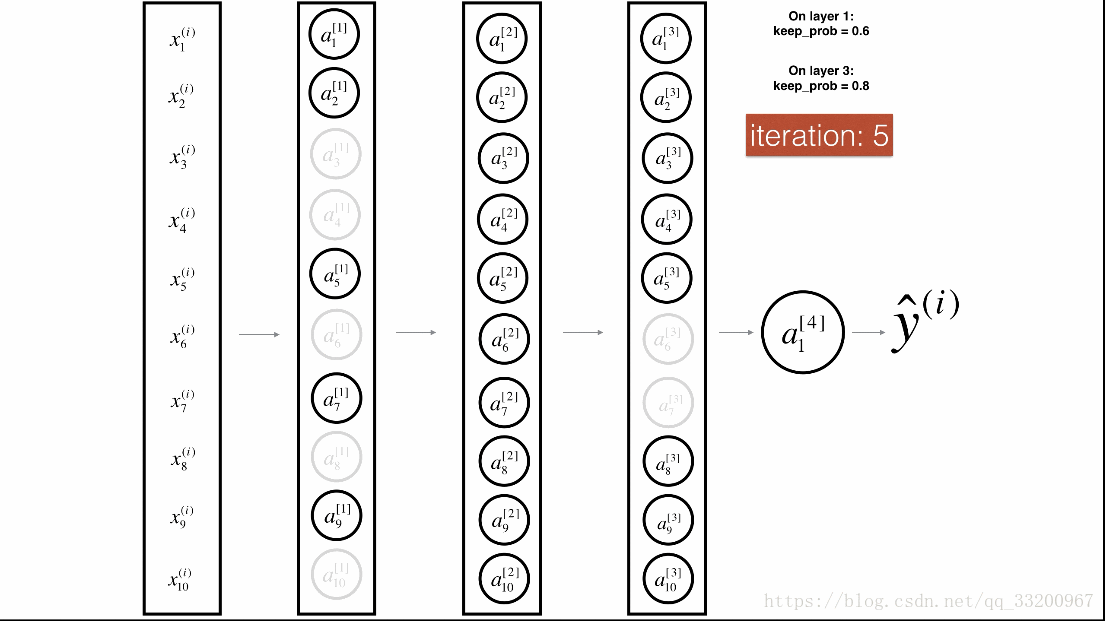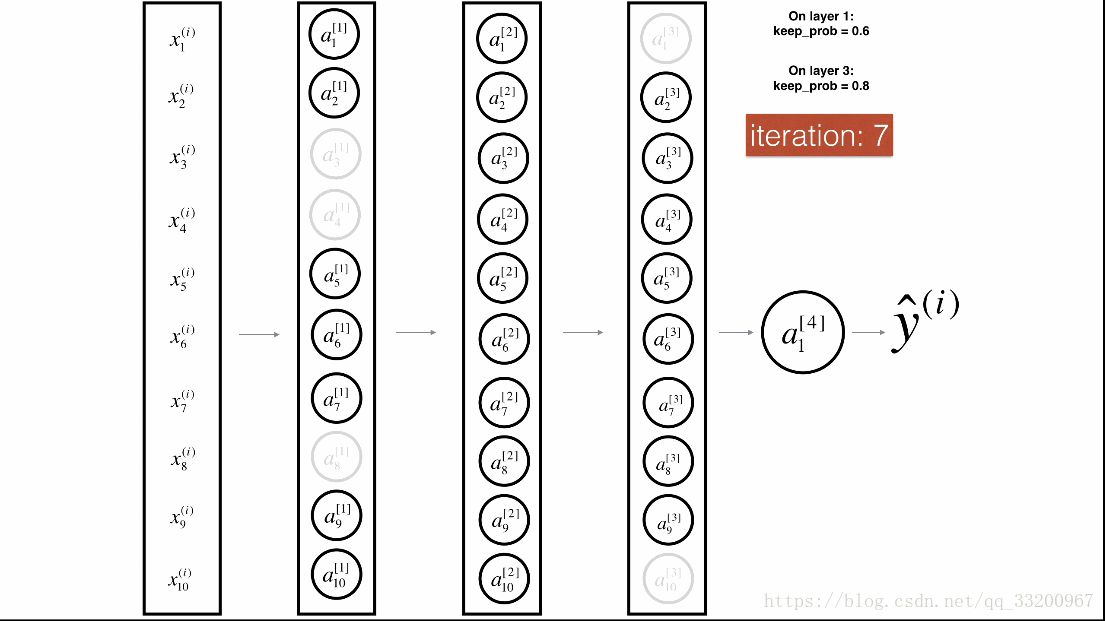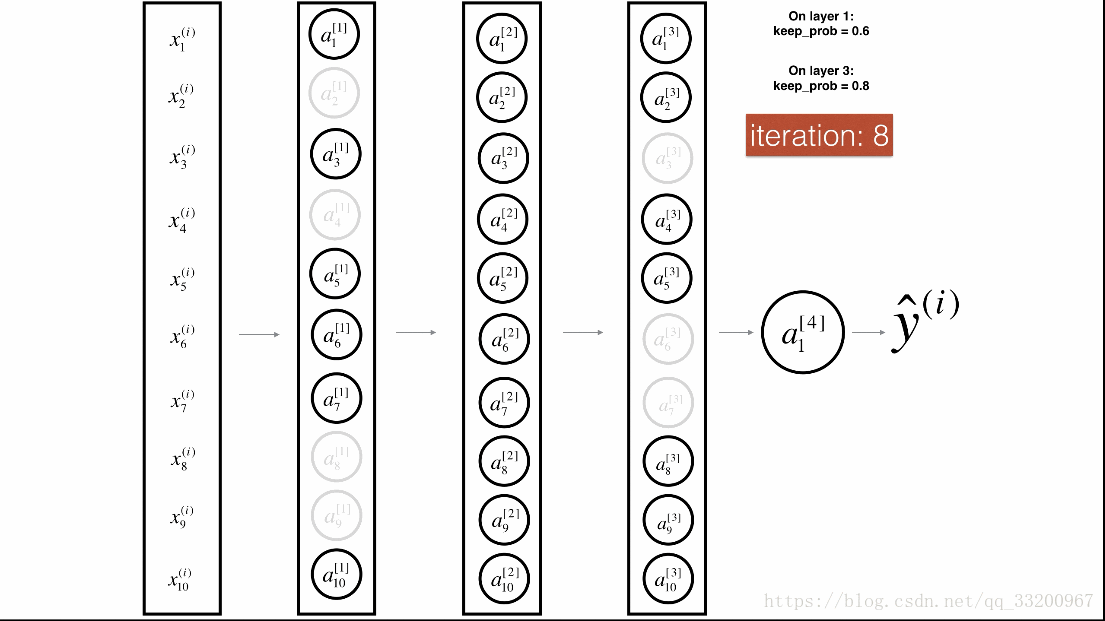
Forward Propagation with Dropout¶
def forward_propagation_with_dropout(X, parameters, keep_prob=0.5):
"""
Implements forward propagation with dropout.
Arguments:
X -- Input dataset, shape (2, number of examples)
parameters -- Dictionary containing parameters W1, b1, W2, b2, W3, b3
keep_prob -- Probability of keeping neurons active
Returns:
A3 -- Output of forward propagation
cache -- Cache for backward propagation
"""
np.random.seed(1)
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
Z1 = np.dot(W1, X) + b1
A1 = relu(Z1)
D1 = np.random.rand(A1.shape[0], A1.shape[1])
D1 = (D1 < keep_prob)
A1 = np.multiply(A1, D1)
A1 = A1 / keep_prob # Scale activation values
Z2 = np.dot(W2, A1) + b2
A2 = relu(Z2)
D2 = np.random.rand(A2.shape[0], A2.shape[1])
D2 = (D2 < keep_prob)
A2 = np.multiply(A2, D2)
A2 = A2 / keep_prob
Z3 = np.dot(W3, A2) + b3
A3 = sigmoid(Z3)
cache = (Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3)
return A3, cache
Backward Propagation with Dropout¶
def backward_propagation_with_dropout(X, Y, cache, keep_prob):
"""
Implements backward propagation with dropout.
Arguments:
X -- Input dataset
Y -- True label vector
cache -- Cache from forward_propagation_with_dropout()
keep_prob -- Probability of keeping neurons active
Returns:
gradients -- Dictionary of gradients
"""
m = X.shape[1]
(Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1. / m * np.dot(dZ3, A2.T)
db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dA2 = np.multiply(dA2, D2) # Apply dropout mask
dA2 = dA2 / keep_prob # Scale gradients
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1. / m * np.dot(dZ2, A1.T)
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
dA1 = np.multiply(dA1, D1) # Apply dropout mask
dA1 = dA1 / keep_prob # Scale gradients
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1. / m * np.dot(dZ1, X.T)
db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3, "dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
Run Dropout Model¶
if __name__ == "__main__":
parameters = model(train_X, train_Y, keep_prob=0.86, learning_rate=0.3)
print("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
Output:
```text
Cost after iteration 0: 0.6543912405149825
Cost after iteration 10000: 0.0610